Mitochondrial-Localized Protein Transcript Abundance can Predict
By A Mystery Man Writer
Last updated 20 May 2024


Constraint-based modeling of yeast mitochondria reveals the

The mitochondrial outer membrane protein MDI promotes local
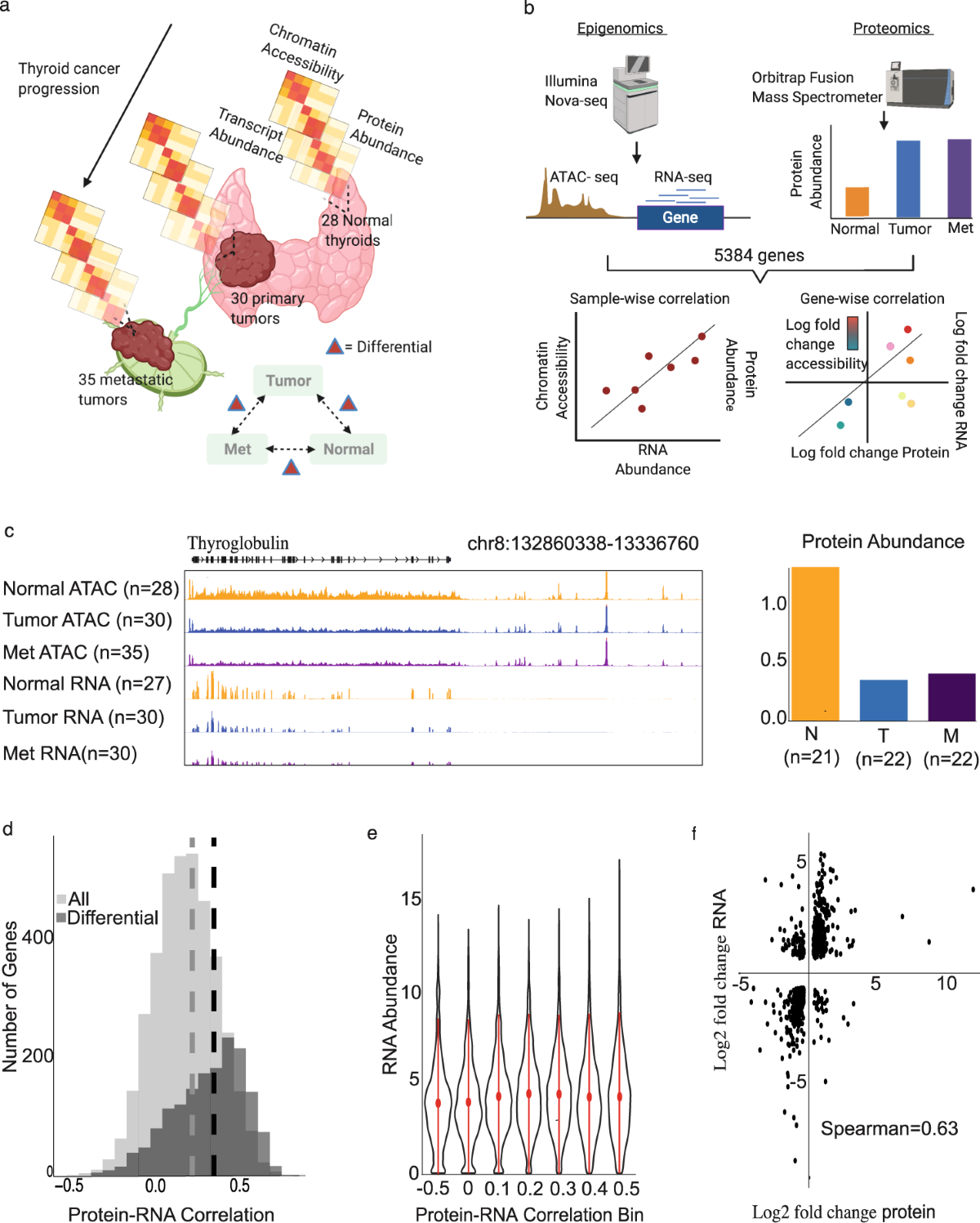
Chromatin accessibility associates with protein-RNA correlation in

Quantifying and Localizing the Mitochondrial Proteome Across Five
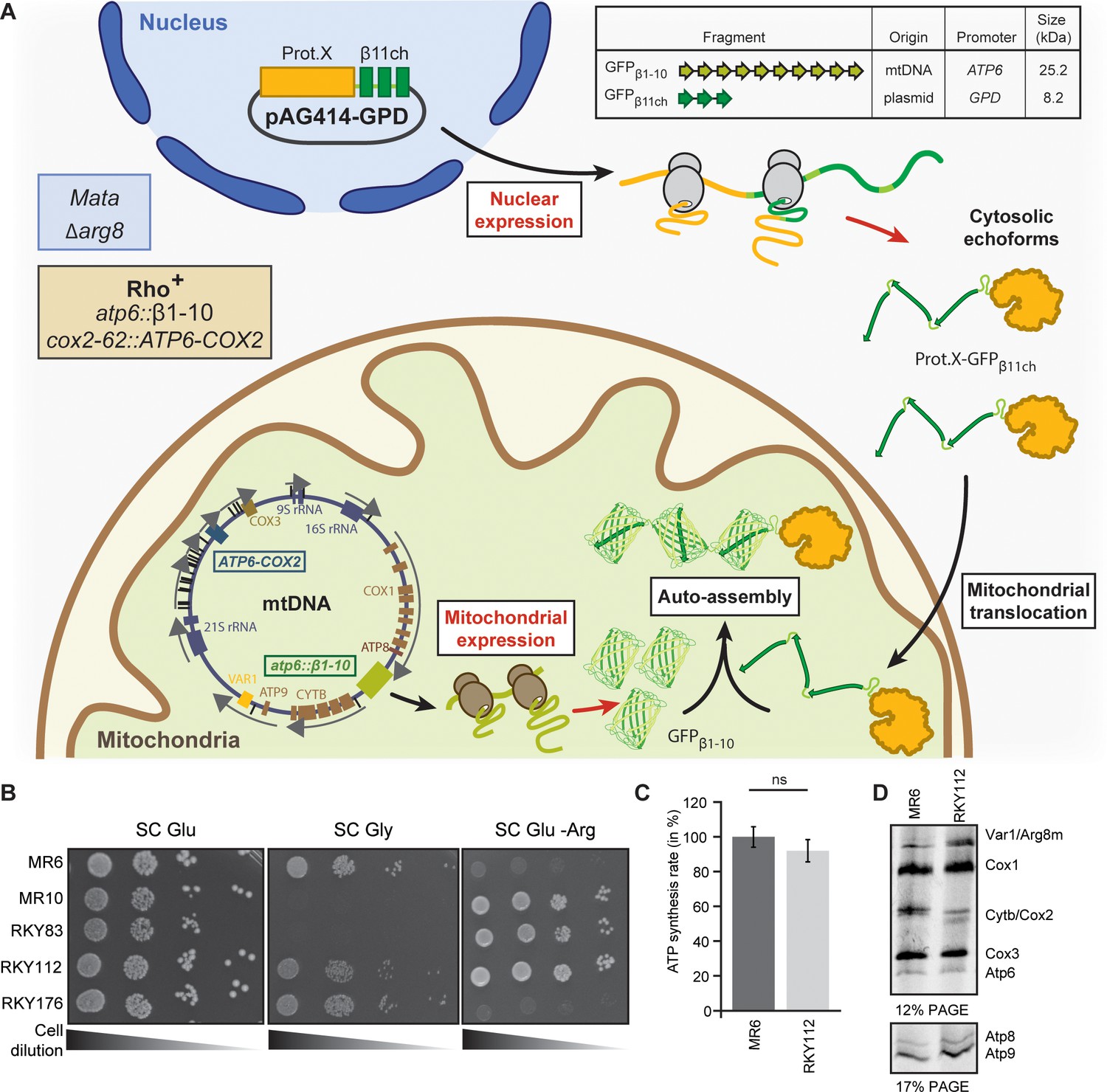
Assigning mitochondrial localization of dual localized proteins
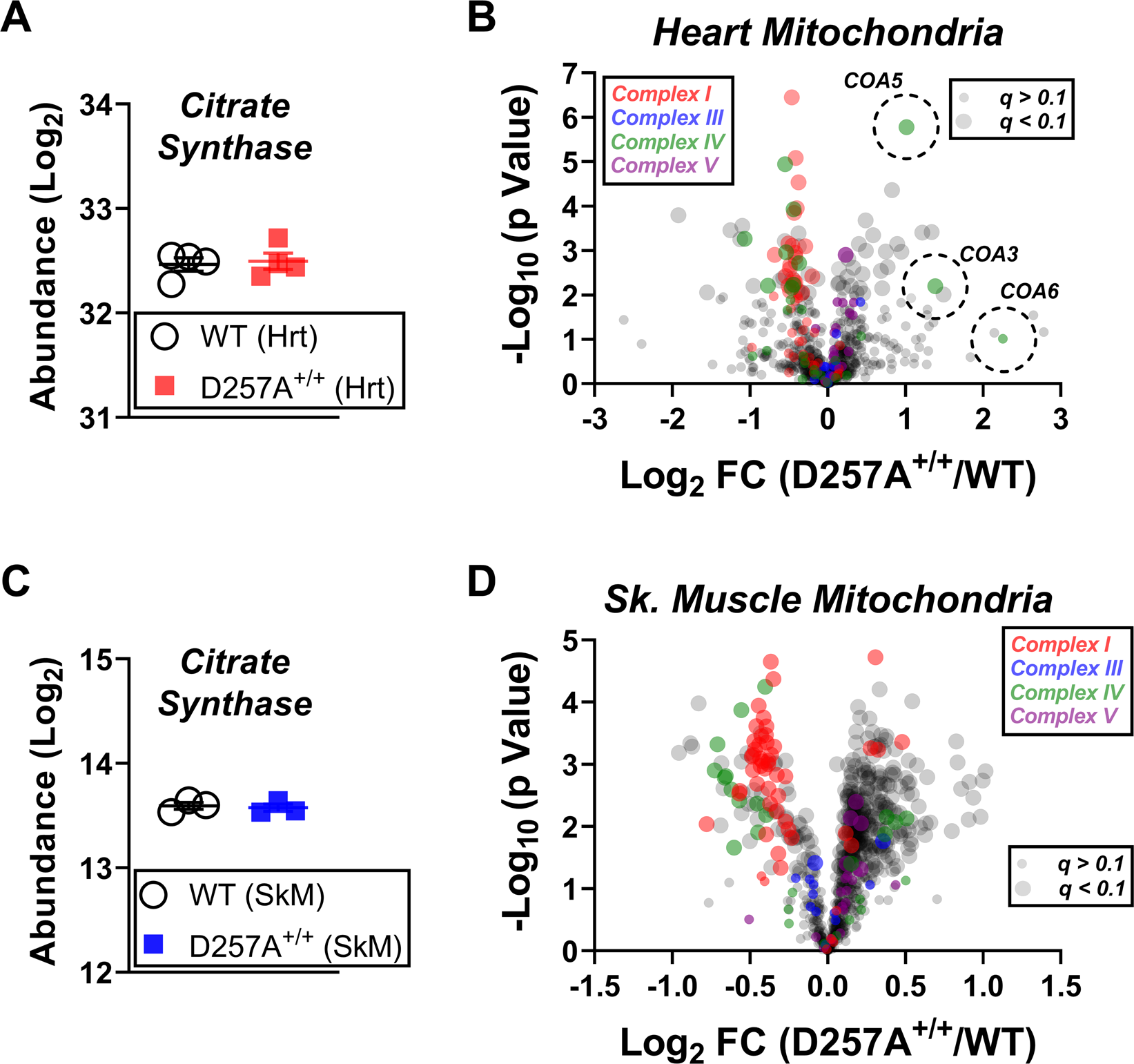
Subcellular proteomics combined with bioenergetic phenotyping
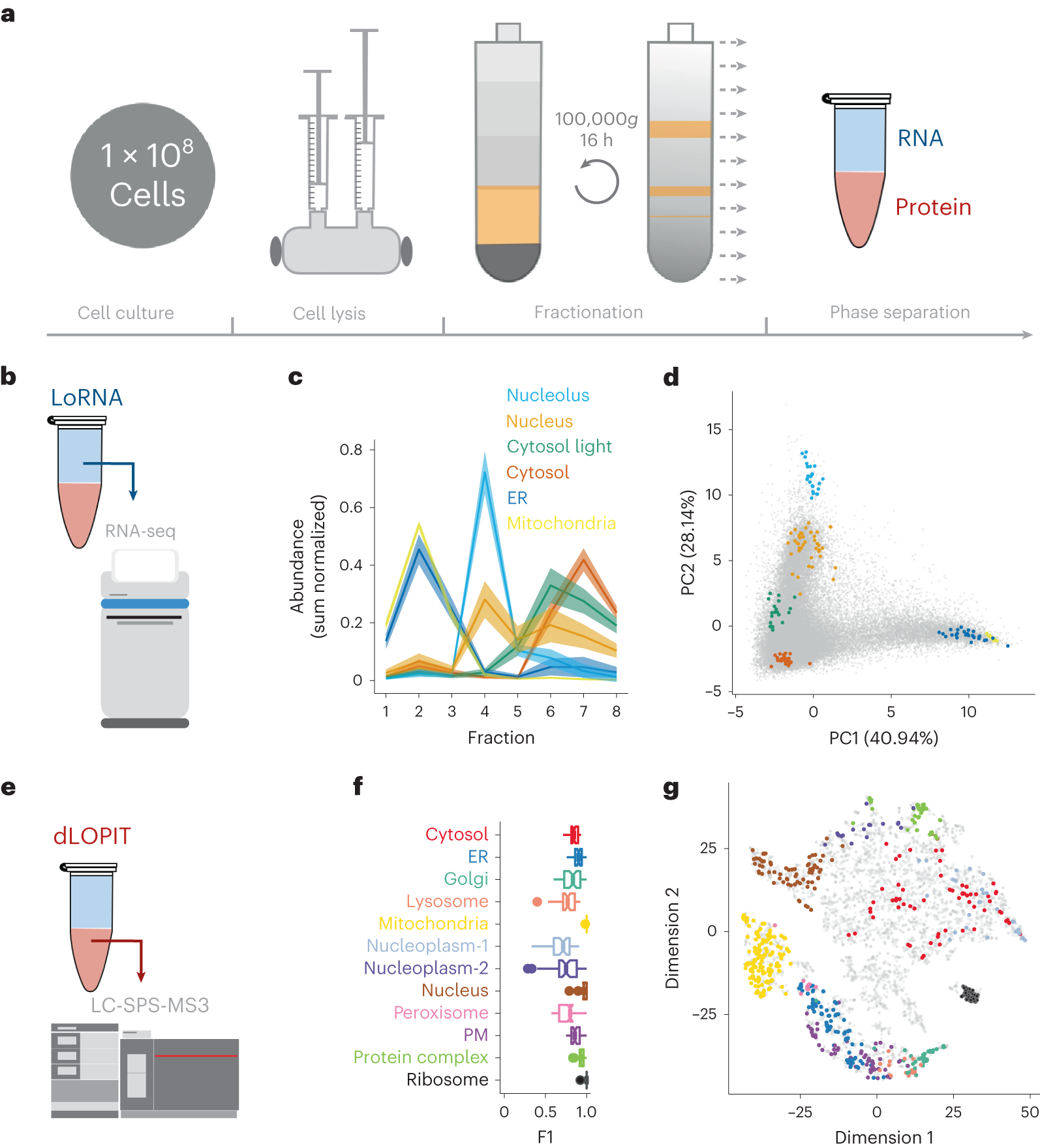
System-wide analysis of RNA and protein subcellular localization

Protein Signatures for Distinguishing Colorectal Cancer Liver Metastases from Primary Liver Cancer Using Tissue Slide Proteomics

Protein prediction models support widespread post-transcriptional
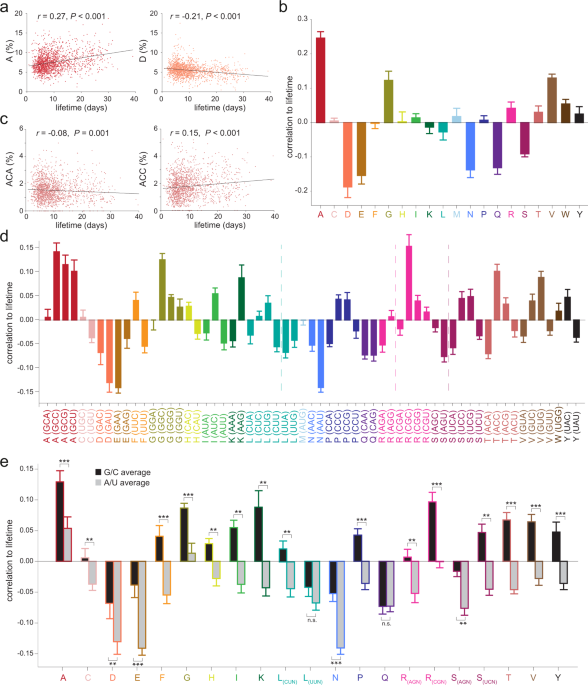
The codon sequences predict protein lifetimes and other parameters
IJMS, Free Full-Text

Figure3 The Expression of Nuclear Genes Encoding Mitochondrial
Protein prediction models support widespread post-transcriptional
Assigning mitochondrial localization of dual localized proteins
Recommended for you
 Congreso Internacional UCEC 202414 Jul 2023
Congreso Internacional UCEC 202414 Jul 2023 UCEC Education – Improving Education Opportunities Together14 Jul 2023
UCEC Education – Improving Education Opportunities Together14 Jul 2023 UCEC 5.25 Inch Front Panel USB Hub with 2-Port USB 3.0 & 2-Port14 Jul 2023
UCEC 5.25 Inch Front Panel USB Hub with 2-Port USB 3.0 & 2-Port14 Jul 2023 UCEC USB 2.0 Video Capture Card Device VHS VCR TV to DVD Converter14 Jul 2023
UCEC USB 2.0 Video Capture Card Device VHS VCR TV to DVD Converter14 Jul 2023- Identification of modules associated with the clinical traits of14 Jul 2023
- UCEC OFFICE - Falun Zhu Trademark Registration14 Jul 2023
 Capture Card 4K-HDR 1080p 60fps With USB-3.0 for PS4 PS5 Xbox14 Jul 2023
Capture Card 4K-HDR 1080p 60fps With USB-3.0 for PS4 PS5 Xbox14 Jul 2023 Identification of the prognostic value of a 2-gene signature of14 Jul 2023
Identification of the prognostic value of a 2-gene signature of14 Jul 2023 UCEC USB 2.0 Video Capture Card Device VHS VCR TV to DVD Converter for Mac OS14 Jul 2023
UCEC USB 2.0 Video Capture Card Device VHS VCR TV to DVD Converter for Mac OS14 Jul 2023 UCEC Left-Angled Micro HDMI to HDMI Male Cable Stretched Length14 Jul 2023
UCEC Left-Angled Micro HDMI to HDMI Male Cable Stretched Length14 Jul 2023
You may also like
 LiveTarget Yellow Perch Rattlebait/ – Pete's Pro14 Jul 2023
LiveTarget Yellow Perch Rattlebait/ – Pete's Pro14 Jul 2023 1998 South Park Kyle Broflovski Fleece Trapper Hat Embroidered Cosplay - collectibles - by owner - sale - craigslist14 Jul 2023
1998 South Park Kyle Broflovski Fleece Trapper Hat Embroidered Cosplay - collectibles - by owner - sale - craigslist14 Jul 2023 A LOOK INSIDE ONE OF MY CATFISH TACKLE BOX14 Jul 2023
A LOOK INSIDE ONE OF MY CATFISH TACKLE BOX14 Jul 2023 Vintage Fishing Caps - CafePress14 Jul 2023
Vintage Fishing Caps - CafePress14 Jul 2023 Braided Fishing Line Westin W10 13 Braid 135 m14 Jul 2023
Braided Fishing Line Westin W10 13 Braid 135 m14 Jul 2023 Japanese Key Hook14 Jul 2023
Japanese Key Hook14 Jul 2023 Zebco Micro 5 ft UL Freshwater Spincast Rod and Reel Combo14 Jul 2023
Zebco Micro 5 ft UL Freshwater Spincast Rod and Reel Combo14 Jul 2023 Las mejores ofertas en Bass Rig Snelled ganchos de pesca14 Jul 2023
Las mejores ofertas en Bass Rig Snelled ganchos de pesca14 Jul 2023 No Fishing between Lines Metal Sign Stock Photo - Image of14 Jul 2023
No Fishing between Lines Metal Sign Stock Photo - Image of14 Jul 2023 From Top to Bottom: A Look at Sinking Lines (Part One) - Anchored Outdoors14 Jul 2023
From Top to Bottom: A Look at Sinking Lines (Part One) - Anchored Outdoors14 Jul 2023